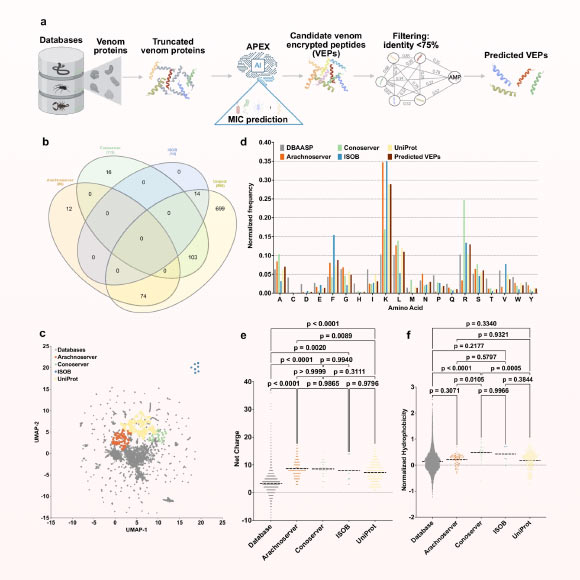
To identify new antimicrobial candidates, scientists at the University of Pennsylvania mined global venomics datasets using a deep-learning system called APEX.
Guan et al. demonstrate that venoms are a rich source of previously hidden antimicrobial scaffolds, and that integrating large-scale computational mining with experimental validation can accelerate the discovery of urgently needed antibiotics. Image credit: Guan et al., doi: 10.1038/s41467-025-60051-6.
The rise of antibiotic-resistant pathogens, particularly gram-negative bacteria, highlights the urgent need for novel therapeutics.
Venoms form an immense and largely untapped reservoir of bioactive molecules with antimicrobial potential.
In the new study, University of Pennsylvania researcher César de la Fuente and his colleagues used the APEX deep-learning system to sift through a database of 16,123 venom proteins and 40,626,260 venom encrypted peptides.
From these, the algorithm identified 386 candidate peptides that are structurally and functionally distinct from known antimicrobial peptides.
“Venoms are evolutionary masterpieces, yet their antimicrobial potential has barely been explored,” Dr. de la Fuente said.
“APEX lets us scan an immense chemical space in just hours and identify peptides with exceptional potential to fight the world’s most stubborn pathogens.”
From the AI-selected shortlist, the scientists synthesized 58 venom peptides for laboratory testing.
Fifty three of them killed drug-resistant bacteria — including Escherichia coli and Staphylococcus aureus — at doses that were harmless to human red blood cells.
“By pairing computational triage with traditional lab experimentation, we delivered one of the most comprehensive investigations of venom derived antibiotics to date,” said Dr. Marcelo Torres, co-author of the study.
“The platform mapped more than 2,000 entirely new antibacterial motifs — short, specific sequences of amino acids within a protein or peptide responsible for their ability to kill or inhibit bacterial growth,” said Dr. Changge Guan, co-author of the study.
“Our team is now taking the top peptide candidates which could lead to new antibiotics and improving them through medicinal-chemistry tweaks.”
The results appear in the journal Nature Communications.
_____
C. Guan et al. 2025. Computational exploration of global venoms for antimicrobial discovery with Venomics artificial intelligence. Nat Commun 16, 6446; doi: 10.1038/s41467-025-60051-6